Mesoscopic Modeling and Rapid Simulation of Incremental Changes in Epidemic Scenarios on GPUs: Fast What–If Analyses of Localized and Dynamic Effects
Abstract
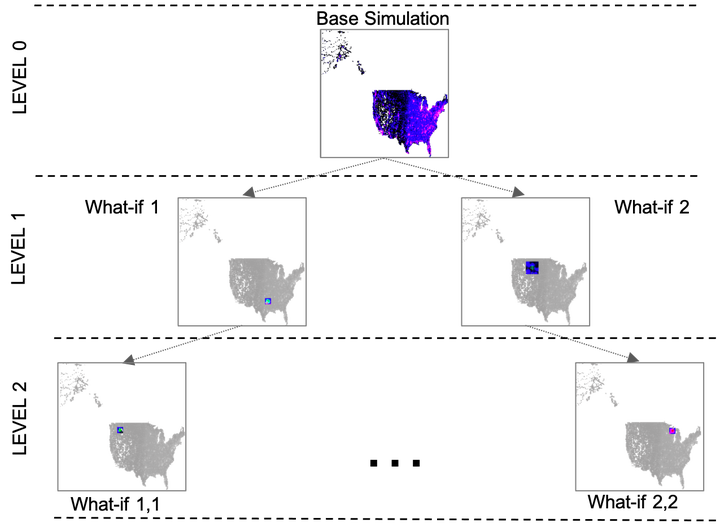
In simulation-based studies and analyses of epidemics, a major challenge lies in resolving the conflict between fidelity of models and the speed of their simulation. Another related challenge arises in dealing with the large number of what–if scenarios that need to be explored. Here, we describe new computational methods that together provide an approach to dealing with both challenges. A mesoscopic modeling approach is described that strikes a middle ground between macroscopic models based on coupled differential equations and microscopic models built on fine-grained behaviors at the individual entity level. The mesoscopic approach offers the ability to incorporate complex compositions of multiple layers of dynamics even while retaining the potential for aggregate behaviors at varying levels. It also is an excellent match to the accelerator-based architectures of modern computing platforms in which graphical processing units (GPUs) can be exploited for fast simulation via the parallel execution mode of single instruction multiple thread (SIMT). The challenge of simulating a large number of scenarios is addressed via a method of sharing model state and computation across a tree of what–if scenarios that are localized, incremental changes to a large base simulation. A combination of the mesoscopic modeling approach and the incremental what–if scenario tree evaluation has been implemented in the software on modern GPUs. Synthetic simulation scenarios are presented to demonstrate the computational characteristics of our approach. Results from the experiments with large population data, including USA, UK, and India, illustrate the modeling methodology and computational performance on thousands of synthetically generated what–if scenarios. Execution of our implementation scaled to 8192 GPUs of supercomputing platforms demonstrates the ability to rapidly evaluate what–if scenarios several orders of magnitude faster than the conventional methods.